h3k27me3 dna methylation
Polycomb Group PcG proteins maintain transcriptional repression throughout development mostly by regulating chromatin structure. DNA methylation is another important layer of epigenetic regulation in addition to histone modifications.

Genomic Profiles Of Dna Methylation Mc And H3k27me3 Expressed As Mean Download Scientific Diagram
Published H3K27me3 ChIP-seq data were from 14 23 and DNA methylation data were from 44 45.

. Mutual antagonism between DNA methylation and H3K27me3 histone methylation suggests a dynamic crosstalk between these epigenetic marks that could help ensure correct gene expression programmes. DNA methylation and H3K27me3 represent two independent epigenetic silencing markers and both can serve as the primary imprints to initiate genomic imprinting. DNA methylation and H3K27me3 are both involved in the establishment and maintenance of epigenetic gene silencing.
Homeobox HOX transcription factors play pivotal roles in many aspects of cellular physiology embryonic. The former was found at hypomethylated regions and the latter at hypermethylated regions of DNA Section 4 suggests maternal H3K27me3 can serve as a methylation-independent imprinting mark and suggests that paternal allele expression of. For DNA methylation tracks gray bars indicate the CpG sites with 5 reads coverage.
H3K27me3 gene expression and DNA methylation in severely DNA hypomethylated mouse somatic cells. It has been reported that DNA methylation can elicit the changes in chromatin structure including histone deacetylation methylation and local chromatin compaction. Accumulating evidence indicates that these two imprinting mechanisms cooperate to.
First higher H3K27me3 levels were confirmed in the genes with the dual modification CNN3 SFRP1 and SLC6A15 than in housekeeping genes EEF1A1. However H3K27me2 shows a similar distribution to H3K27 while H3K27me1 is associated. We suggest that through facilitating correct PRC2-target-ing an intact DNA methylome is required for appropriate.
Histone H3K27 Modifications When H3K27 is trimethylated it is tightly associated with inactive gene promotersThe mono- and di-methylation states are less studied. Some evidence points toward a cooperative relationship. H3K27me3 is an epigenetic modification to the DNA packaging protein Histone H3It is a mark that indicates the tri-methylation of lysine 27 on histone H3 proteinThis tri-methylation is associated with the downregulation of nearby genes via the formation of.
Additionally the balance between the transcriptionally favorable tri-methylation histone H3 at lysine 4 H3K4me3 and the unfavorable tri-methylation histone H3 at lysine 27 H3K27me3 is shifted in the Dicer-deficient epigenome. Faithful maintenance of genomic imprinting is essential for mammalian development. Section 3 suggested that H3K27me3 and H3K9me3 are involved in rendering maternal alleles inaccessible.
One of them the trimethylation of histone H3 lysine 27 H3K27me3 confers a repressive chromatin state with gene silencing. Silencing of developmental genes by H3K27me3 and DNA methylation reflects the discrepant plasticity of embryonic and extraembryonic lineages Cell Res. D Sequence preference of H3K27me3 and DNAme for different CpG content.
Our results uncover an unexpected relationship between DNA methylation and transcriptional repression by Polycomb. For example the polycomb group protein EZH2 has been shown to positively regulate DNA methylation. Epigenetic regulation controls multiple aspects of the plant development.
Work from Manzo et al 2017 now shows that an isoform of de novo DNA methyltransferase DNMT3A provides specificity in the system by. As a consequence this may also enhance the reduction of H3K27me3 at normal PRC2 targets due to dilution of PRC2 molecules. H3K9me3 and H3K27me3 are two known histone marks that seem to promote chromatin compaction in promoter regions which could be associated with hindrance of transcriptional.
The Myh6 and Nppa promoters have been previously shown to be demethylated in αMHC-GFP iCMs at week 4 Ieda et al 2010. Mapping epigenetic marks in severely DNA hypomethylated Dnmt1--mouse somatic cells. H3K27me3 Profiling of the Endosperm Implies Exclusion of Polycomb Group Protein Targeting by DNA Methylation Isabelle Weinhofer1 Elisabeth Hehenberger1 Pawel Roszak1 Lars Hennig12 Claudia Kohler12 1Department of Biology and Zurich-Basel Plant Science Center Swiss Federal Institute of Technology Zurich Switzerland 2Department of Plant Biology and Forest.
There are data showing coordinate regulation between the marks. As a first step to investigate the interplay between DNA methylation and PRC2 in the context of gene regulation we generated maps of DNA methylation H3K4me3 and the PRC2-signature H3K27me3 histone mark in cells where DNA methylation is strongly reduced. The existence of DNA methylation and H3K27me3 on the same DNA molecules was analyzed in two cancer cell lines PC3 and MCF7 by bisulfite sequencing of chromatin immunoprecipitated DNA 32 41.
While germline DNA methylation-dependent canonical imprinting is relatively stable during development the recently found oocyte-derived H3K27me3-mediated noncanonical imprinting is mostly transient in early embryos with some genes important for placental development. These findings are consistent with our results with one notable. We show that DNA methylation profiling distinguishes MPNST from its histological mimics was unrelated to anatomical site and formed two main clusters MeGroups 4 and 5.
The dynamics of H3K27me3-dependent imprinting is strikingly different from DNA methylation-dependent imprinting that is largely maintained in both embryonic and extra-embryonic lineages 36. Therefore we investigated whether and how DNA methylation re-patterns at cardiac promoters during iCM reprogramming. The N-terminal tail of histone can be differently modified to regulate various chromatin activities.
The published data of H3K27me3 enrichment 1 and DNA methylation 2 in pre-implantation embryos were used in these analyses. H3K27me3 is also an important mark of the inactive X chromosome Xi Rougeulle et al 2004. Polycomb Repressive Complex 2 PRC2 a component of the Polycomb machinery is responsible for the methylation of histone.
C and D Representative genome browser views of an oocyte DNA methylationdependent PEG Snrpn C and a maternal H3K27me3dependent PEG Gab1 D. Two studies in which epigenetic silencing marks were profiled in prostate cancer cells PC3 revealed that loci marked with H3K27me3 are devoid of DNA hypermethylation raising the possibility that gene silencing by H3K27me3 occurs independently of promoter methylation 2021. H3K27me3 loss is rarely seen in other highgrade sarcomas and was not found to be associated with an inferior outcome in MPNST.
Global DNA hypomethylation may allow for increased binding of PRC2 and establishment of H3K27me3 at genomic sites that are normally protected by the DNA methylation mark.
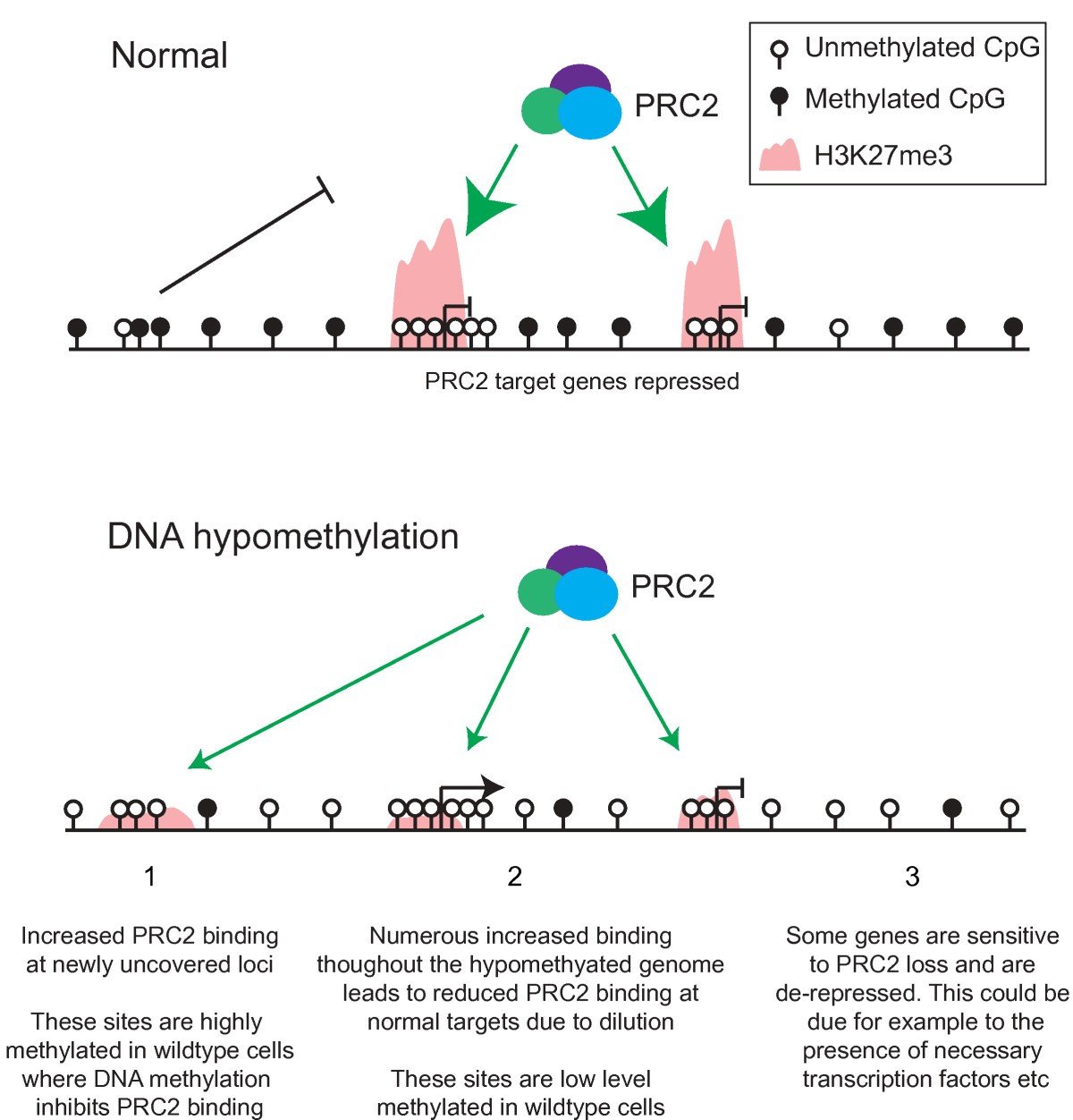
Redistribution Of H3k27me3 Upon Dna Hypomethylation Results In De Repression Of Polycomb Target Genes Genome Biology Full Text

Somatic Maintenance Of Oocyte Derived Dna Methylation And H3 Lysine 27 Download Scientific Diagram
Plos One Relationship Between Gene Body Dna Methylation And Intragenic H3k9me3 And H3k36me3 Chromatin Marks

H3k9me3 H3k27me3 And Dna Methylation At Ervs In E13 5 Pgcs A Download Scientific Diagram

Reduction In Dna Methylation Affected The Distribution Of H3k27me3 But Download Scientific Diagram

Dna Methylation And Histone Mark Patterns In Oocyte A At Day 5 Download Scientific Diagram

Reversible Regulation Of Promoter And Enhancer Histone Landscape By Dna Methylation In Mouse Embryonic Stem Cells Cell Reports

Genome Wide Identification Of H3k27me3 Regions A Comparison Of Download Scientific Diagram

Cross Talk Between Dna Methylation Histone Modifications And Download Scientific Diagram

Enrichment Of H3k9me3 H3k27me3 And Gene Expression In E13 5 Pgcs A Download Scientific Diagram

Dna Methylation And H3k4me3 H3k27me3 Are Mutually Exclusive On The Download Scientific Diagram
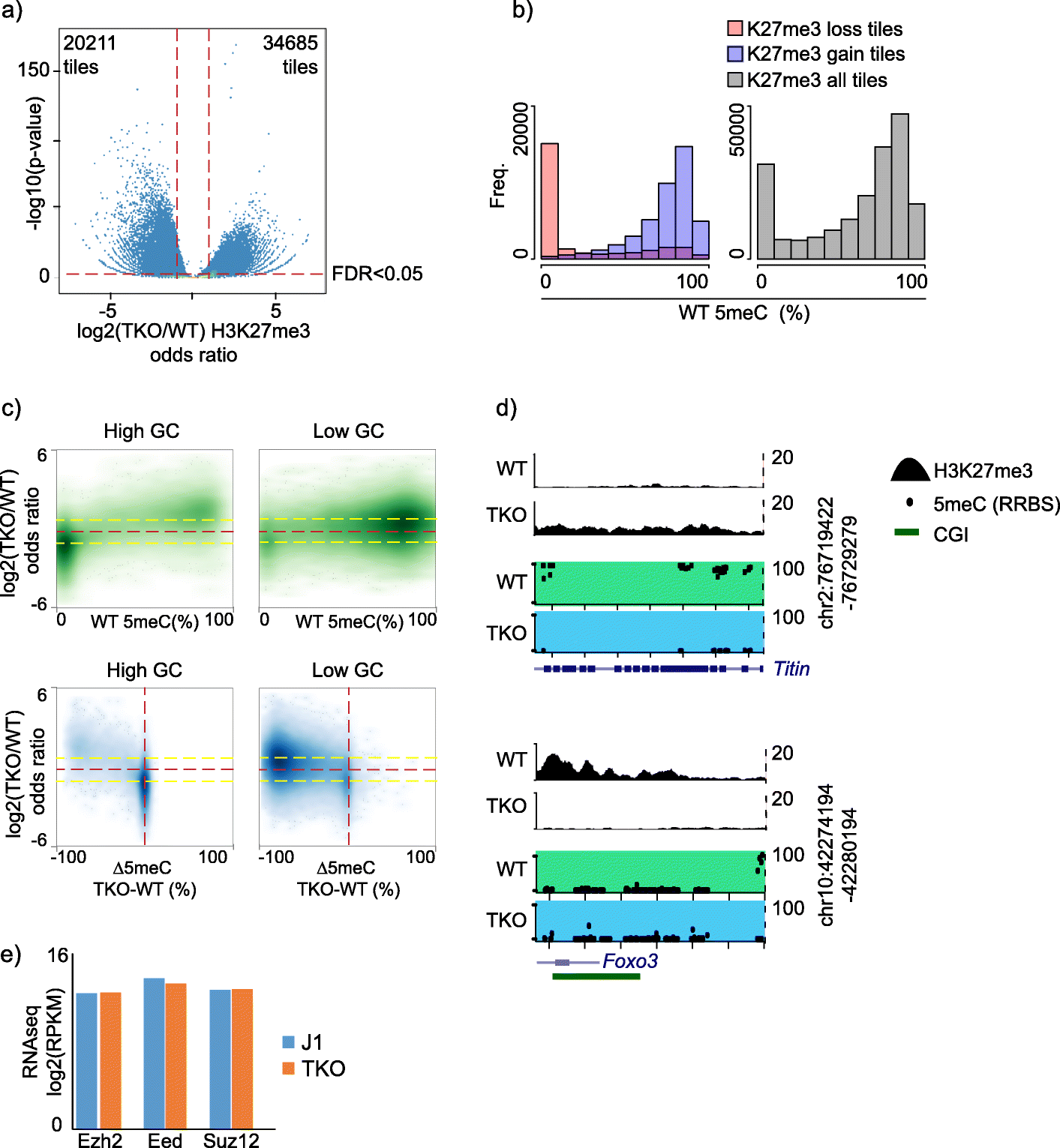
Bivalent Promoter Hypermethylation In Cancer Is Linked To The H327me3 H3k4me3 Ratio In Embryonic Stem Cells Bmc Biology Full Text

The Crosstalk Between Prc2 Nsd1 And Dnmt3a On Chromatin A Download Scientific Diagram

Model For How H3k27me3 And H2ak119ub Epigenetic Signatures Modulate Dna Download Scientific Diagram

H3k27me3 Enriched Cpg Island Promoters Are Devoid Of Dna Methylation Download Scientific Diagram
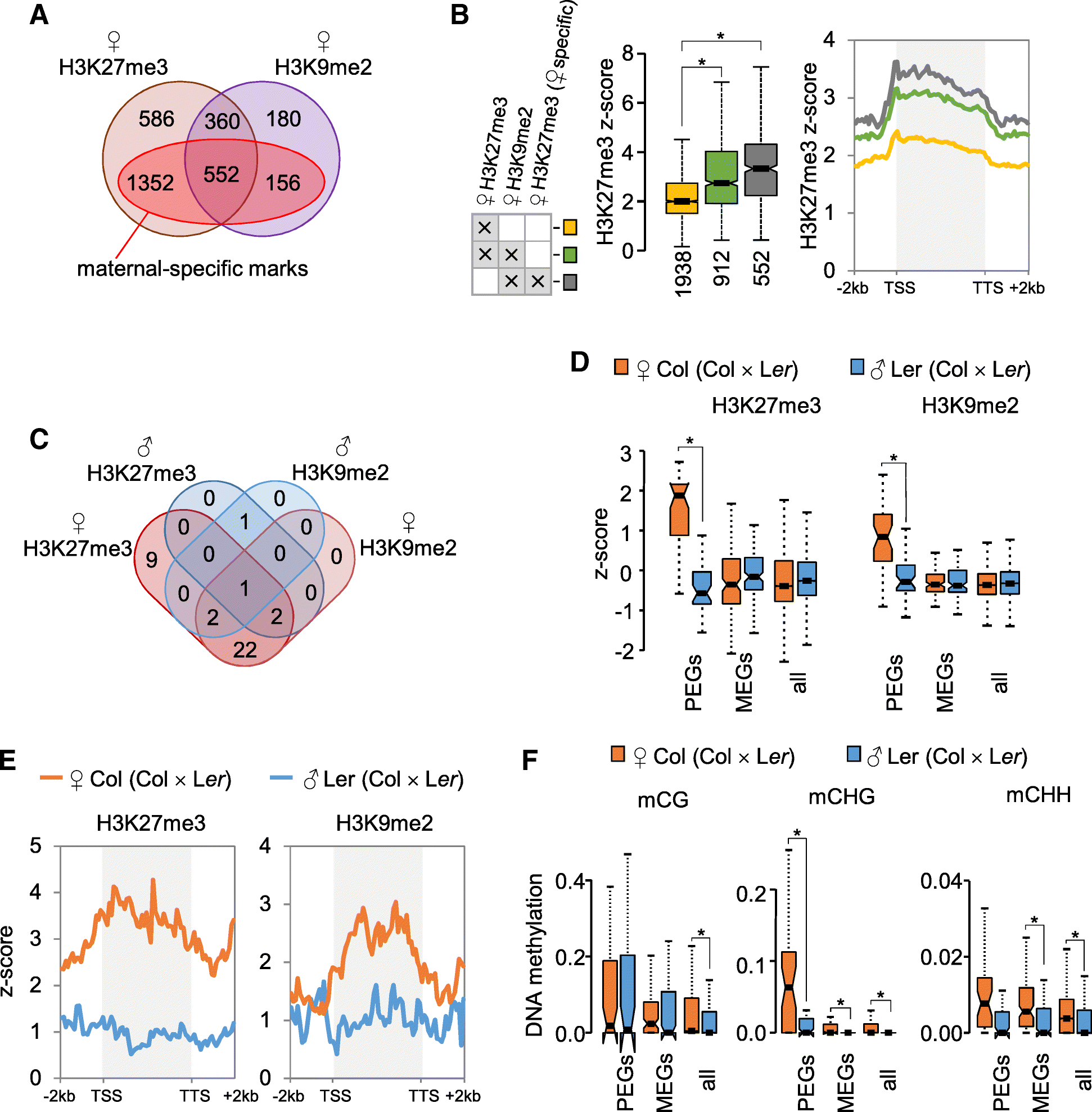
Epigenetic Signatures Associated With Imprinted Paternally Expressed Genes In The Arabidopsis Endosperm Genome Biology Full Text

Bischip Seq Dna Methylation Profiles Of H3k27me3 Enriched Dna From Download Scientific Diagram

H3k27me3 And H3k4me3 Mapping At Gene Promoters In Dna Hypomethylated Download Scientific Diagram

Crosstalk Of H3k36 Methylation With H3k27 Methylation And Dna Download Scientific Diagram
Comments
Post a Comment